Nano-Positioning System (NPS-Software)
2017
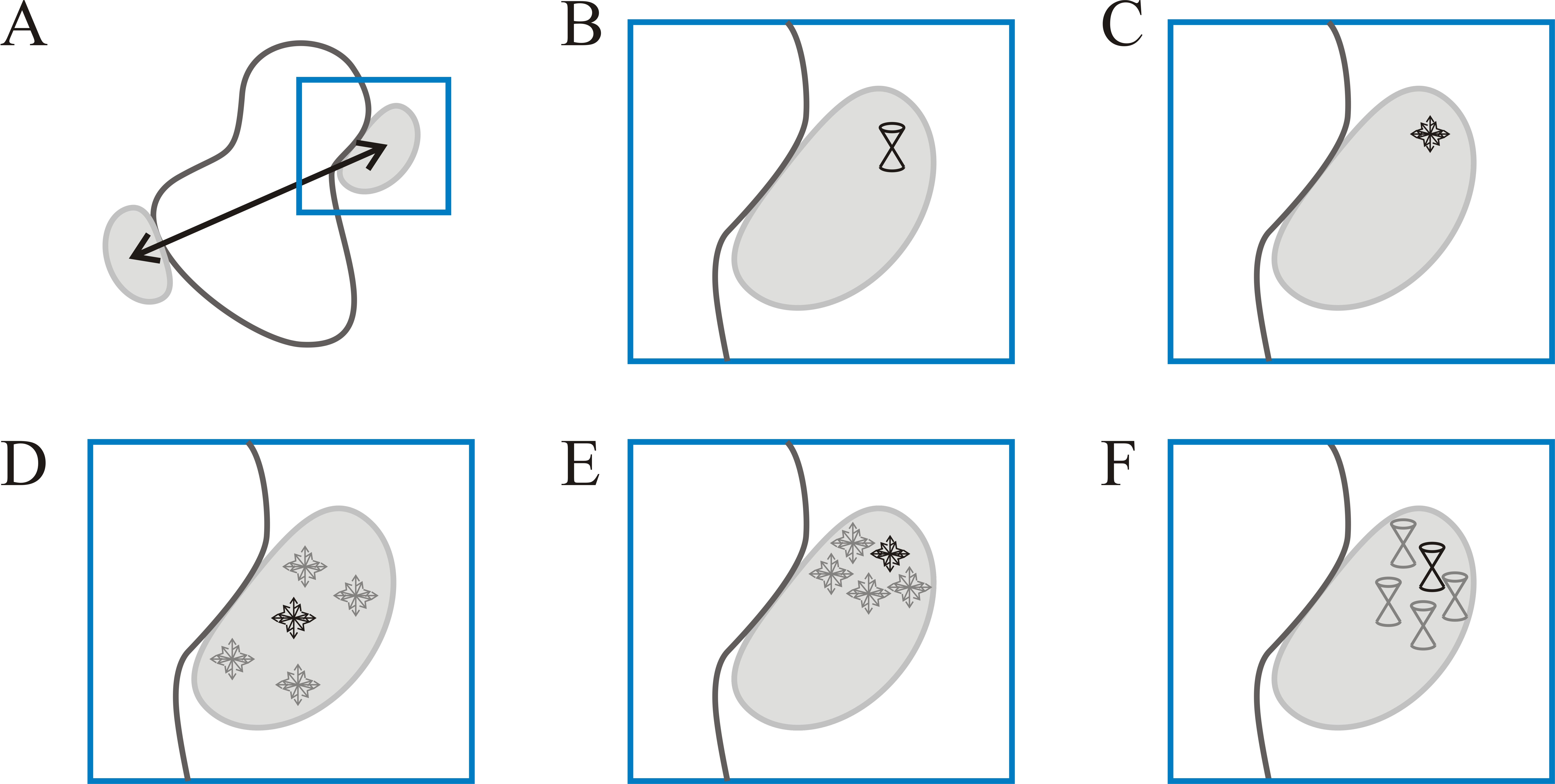
Here we present a detailed protocol for obtaining smFRET data and applying the Fast-NPS. We provide detailed instructions for the acquisition of the three input parameters of Fast-NPS: the smFRET values, as well as the quantum yield and anisotropy of the dye molecules. Recently, the NPS has been used to elucidate the architecture of an archaeal open promotor complex. This data is used to demonstrate the influence of the five different dye models on the posterior distribution.
Thilo Dörfler, Tobias Eilert, Carlheinz Röcker, Julia Nagy, Jens Michaelis
Structural Information from Single-molecule FRET Experiments Using the Fast Nano-positioning System
Single-molecule Förster Resonance Energy Transfer (smFRET) can be used to obtain structural information on biomolecular complexes in realtime. Thereby, multiple smFRET measurements are used to localize an unknown dye position inside a protein complex by means of trilateration. In order to obtain quantitative information, the Nano-Positioning System (NPS) uses probabilistic data analysis to combine structural information from X-ray crystallography with single-molecule fluorescence data to calculate not only the most probable position but the complete threedimensional probability distribution, termed posterior, which indicates the experimental uncertainty. The concept was generalized for the analysis of smFRET networks containing numerous dye molecules. The latest version of NPS, Fast-NPS, features a new algorithm using Bayesian parameter estimation based on Markov Chain Monte Carlo sampling and parallel tempering that allows for the analysis of large smFRET networks in a comparably short time. Moreover, Fast-NPS allows the calculation of the posterior by choosing one of five different models for each dye, that account for the different spatial and orientational behavior exhibited by the dye molecules due to their local environment.
Journal of Visualized Experiments, 2017, 120
doi:10.3791/54782
2015
We further optimized our software by replacing the nested sampler with a custom replica exchange Markov Chain Monte Carlo algorithm. This immensely reduce the calculation time and furthermore enables the use of more features (i.e. applying constraints).
For further information see our latest publication in Faraday Discussions.
M. Beckers, F. Drechsler, T. Eilert, J. Nagy and J. Michaelis*
Quantitative structural information from single-molecule FRET
Single-molecule studies can be used to study biological processes directly and in realtime. In particular, the fluorescence energy transfer between reporter dye moleculesattached to specific sites on macromolecular complexes can be used to infer distance information. When several measurements are combined, the information can be usedto determine the position and conformation of certain domains with respect to the complex. However, data analysis schemes that include all experimental uncertaintiesare highly complex, and the outcome depends on assumptions about the state of the dye molecules. Here, we present a new analysis algorithm using Bayesian parameterestimation based on Markov Chain Monte Carlo sampling and parallel tempering termed Fast-NPS that can analyse large smFRET networks in a relatively short timeand yields the position of the dye molecules together with their respective uncertainties. Moreover, we show what effects different assumptions about the dyemolecules have on the outcome. We discuss the possibilities and pitfalls in structure determination based on smFRET using experimental data for an archaeal transcriptionpre-initiation complex, whose architecture has recently been unravelled by smFRET measurements.
Faraday Discuss., 2015, 184, 117-129
DOI: 10.1039/c5fd00110b
2011
A newer software with advanced capabilities is available upon request. For more details see our publication in the Journal of Physical Chemistry B. For a short comparison of the two software versions please see the following PDF: software comparison
A. Muschielok, J. Michaelis
Application of the Nano-Positioning System to the Analysis of Fluorescence Resonance Energy Transfer Networks
Single-molecule fluorescence resonance energy transfer (sm-FRET) has been recently applied to distance and position estimation in macromolecular complexes. Here, we generalize the previously published Nano-Positioning System (NPS), a probabilistic method to analyze data obtained in such experiments, which accounts for effects of restricted rotational freedom of fluorescent dyes, as well as for limited knowledge of the exact dye positions due to attachment via flexible linkers. In particular we show that global data analysis of complete FRET networks is beneficial and that the measurement of FRET anisotropies in addition to FRET efficiencies can be used to determine accurately both position and orientation of the dyes. This measurement scheme improves localization accuracy substantially, and we can show that the improvement is a consequence of the more precise information about the transition dipole moment orientation of the dyes obtained by FRET anisotropy measurements. We discuss also rigid body docking of different macromolecules by means of NPS, which can be used to study the structure of macromolecular complexes. Finally, we combine our approach with common FRET analysis methods to determine the number of states of a macromolecule.
Journal physical chemistry B 115 (2011) 11927-11937
Supplementary Material
2008
We have recently developed a new method, termed Nano-Positioning System (NPS) which allows the localisation of flexible domains in macromolecules. The method is based on single molecule FRET measurements and a structural model for the core of the macromolecule.
The method is published and explained in Nature Methods.
A. Muschielok, J. Andrecka, A. Jawhari, F. Brückner, P. Cramer, and J. Michaelis
A nano-positioning system for macromolecular structural analysis
By combining single pair FRET data, x-ray crystallographical data and statistical data analysis we have developed a novel method for determining the position of a flexible domain inside of a macromolecule/macromolecular complex. The method shares some similarities to GPS, the global positioning system, which is why we called it NPS, Nano Positioning System.
Nature Methods, 5 (2008 ) 965-971
You can download here a user guide which gives a step by step description on how to use the NPS software. The software requires a Matlab runtime to be installed on the computer. If you do not have Matlab, please write an email to Michaelis and we will send you a software that can be run as a standalone software.
The entire software is based on graphical user interfaces and should therefore be easy to use and comprehend. If you have any questions or comments about the software or would like to see the source code just send an email. If you use the software for your own data analysis please cite the Nature Methods paper.
Note: Due to Mathworks licensing policies we can only provide the necessary Matlab runtime upon request. If you do not have access to Matlab runtime please contact: Jens Michaelis
