Publications AG Gebhardt
2025
Increasingly efficient chromatin binding of cohesin and CTCF supports chromatin architecture formation during zebrafish embryogenesis
J. Coßmann, P.I. Kos, V. Varamogianni-Mamatsi, D. Assenheimer, T. Bischof, T. Kuhn, T. Vomhof, A. Papantonis, L. Giorgetti, and J.C.M. Gebhardt
Nature Communications 16, 1833 (2025), https://doi.org/10.1038/s41467-025-56889-5
Mechanisms of gene regulation by SRCAP and H2A.Z
A. Tollenaere, E. Ugur*, S. Dalla-Longa*, C. Deluz, D. Assenheimer, J.C.M. Gebhardt, H. Leonhardt, D.M. Suter
bioRxiv (2025), (*equal contribution) https://doi.org/10.1101/2024.06.07.597899
Effective in vivo binding energy landscape illustrates kinetic stability of RBPJ-DNA binding
D. Huynh*, P. Hoffmeister*, T. Friedrich, K. Zhang, M. Bartkuhn, F. Ferrante, B.D. Giaimo, R. Kovall, T. Borggrefe, F. Oswald#, and J.C.M. Gebhardt#
Nature Communications 16, 1259 (2025), (*equal contribution, #corresponding author) https://doi.org/10.1038/s41467-025-56515-4
2023
Transcriptional reprogramming by mutated IRF4 in lymphoma
N. Schleussner*, P. Cauchy*, V. Franke, M. Giefing, O. Fornes, N. Vankadari, S.A. Assi, M. Costanza, M.A. Weniger, A. Akalin, I. Anagnostopoulos, T. Bukur, M.G. Casarotto, F. Damm, O. Daumke, B. Edginton-White, J.C.M. Gebhardt, M. Grau, S. Grunwald, M.-L. Hansmann, S. Hartmann, L. Huber, E. Kärgel, S. Lusatis, D. Noerenberg, N. Obier, U. Pannicke, A. Fischer, A. Reisser, A. Rosenwald, K. Schwarz, S. Sundararaj, A. Weilemann, W. Winkler, W. Xu, G. Lenz, K. Rajewsky, W.W. Wasserman, P.N. Cockerill, C. Scheidereit, R. Siebert, R. Küppers, R. Grosschedl, M. Janz, C. Bonifer, and S. Mathas
Nature Communications 14, 6947 (2023), (*equal contribution) https://doi.org/10.1038/s41467-023-41954-8
A multimorphic mutation in IRF4 causes human autosomal dominant combined immunodeficiency
IRF4 International Consortium, O. Fornes, A. Jia, H.S. Kuehn, Q. Min, U. Pannicke, N. Schleussner, R. Thouenon, Z. Yu, M. De Los Angeles Astbury, C.M. Biggs, M. Galicchio, J.A. Garcia-Campos, S. Gismondi, G. Gonzalez Villarreal, K.J. Hildebrand, M. Hönig, J. Hou, D. Moshous, S. Pittaluga, X. Qian, J. Rozmus, A.S. Schulz, A.T. Staines-Boone, B. Sun, J. Sun, S. Uwe, E. Venegas-Montoya, W. Wang, X. Wang, W. Ying, X. Zhai, Q. Zhou, A. Akalin, I. André, T.F.E. Barth, B. Baumann, A. Brüstle, G. Burgio, J.C. Bustamante, J.-L. Casanova, M.G. Casarotto, M. Cavazzana, L. Chentout, I.A. Cockburn, M. Costanza, C. Cui, O. Daumke, K.L. Del Bel, H. Eibel, X. Feng, V. Franke, J.C.M. Gebhardt, A. Götz, S. Grunwald, B. Hoareau, T.R. Hughes, E.-M. Jacobsen, M. Janz, A. Jolma, C. Lagresle-Peyrou, N. Lai, Y. Li, S. Lin, H.Y. Lu, S.O. Lugo-Reyes, X. Meng, P. Möller, N. Moreno-Corona, J.E. Niemela, G. Novakovsky, J.J. Perez-Caraballo, C. Picard, L. Poggi, M.-E. Puig-Lombardi, K.L. Randall, A. Reisser, Y. Schmitt, S. Seneviratne, M. Sharma, J. Stoddard, S. Sundararaj, H. Sutton, L.Q. Tran, Y. Wang, W.W. Wasserman, Z. Wen, W. Winkler, E. Xiong, A.W.H. Yang, M. Yu, L. Zhang, H. Zhang, Q. Zhao, X. Zhen, A. Enders, S. Kracker, R. Martinez-Barricarte, S. Mathas, S.D. Rosenzweig, K. Schwarz, S.E. Turvey, and J.-Y. Wang
Science Immunology 8, eade7953 (2023), https://doi.org/10.1126/sciimmunol.ade7953
2022
Single-molecule tracking of Nodal and Lefty in live zebrafish embryos supports hindered diffusion model
T. Kuhn, A.N. Landge*, D. Mörsdorf*, J. Coßmann, J. Gerstenecker, D. Čapek, P. Müller#, and J.C.M. Gebhardt#
Nature Communications 13, 6101 (2022), (*equal contribution, #corresponding author) https://doi.org/10.1038/s41467-022-33704-z
Stress induced TDP-43 mobility loss independent of stress granules
L. Streit, T. Kuhn, T. Vomhof, V. Bopp, A.C. Ludolph, J.H. Weishaupt, J.C.M. Gebhardt, J. Michaelis#, and K.M. Danzer#
Nature Communications 13, 5480 (2022), (#corresponding author) https://doi.org/10.1038/s41467-022-32939-0
Quantum Optical Microphone in the Audio Band
R. Nold, C. Babin, J. Schmidt, T. Linkewitz, M.T.P. Zaballos, R. Stöhr, R. Kolesov, V. Vorobyov, D.M. Lukin, R. Boppert, S. Barz, J. Vučković, J.C.M. Gebhardt, F. Kaiser, and J. Wrachtrup
PRX Quantum 3, 020358 (2022), https://doi.org/10.1103/PRXQuantum.3.020358
A guide to changing paradigms of glucocorticoid receptor function—a model system for genome regulation and physiology
S. Vettorazzi, D. Nalbantoglu, J.C.M. Gebhardt, and J. Tuckermann
The FEBS Journal 289, 5718–5743 (2022), https://doi.org/10.1111/febs.16100
Single-molecule tracking (SMT) and localization of SRF and MRTF transcription factors during neuronal stimulation and differentiation
O. Kuchler, J. Gerlach, T. Vomhof, J. Hettich, J. Steinmetz, J.C.M. Gebhardt, J. Michaelis#, and B. Knöll#
Open Biology 12, 210383 (2022), (#corresponding author) https://doi.org/10.1098/rsob.210383
Myosin VI regulates the spatial organisation of mammalian transcription initiation
Y. Hari-Gupta, N. Fili, Á. Dos Santos, A.W. Cook, R.E. Gough, H.C.W. Reed, L. Wang, J. Aaron, T. Venit, E. Wait, A. Grosse-Berkenbusch, J.C.M. Gebhardt, P. Percipalle, T.-L. Chew, M. Martin-Fernandez, and C.P. Toseland
Nature Communications 13, 1346 (2022), https://doi.org/10.1038/s41467-022-28962-w
Periodic synchronization of isolated network elements facilitates simulating and inferring gene regulatory networks including stochastic molecular kinetics
J. Hettich and J.C.M. Gebhardt
BMC Bioinformatics 23, 1–30 (2022), https://doi.org/10.1186/s12859-021-04541-6
2021
Transcription Factor RBPJL Is Able to Repress Notch Target Gene Expression but Is Non-Responsive to Notch Activation
L. Pan*, P. Hoffmeister*, A. Turkiewicz, N.N.D. Huynh, A. Große-Berkenbusch, U. Knippschild, J.C.M. Gebhardt, B. Baumann, T. Borggrefe#, and F. Oswald#
Cancers 13, 5027 (2021), (*equal contribution, #corresponding author) https://doi.org/10.3390/cancers13195027
A Fibrinogen Alpha Fragment Mitigates Chemotherapy-Induced MLL Rearrangements
J. Eberle, R.S. Wiehe, B. Gole, L.J. Mattis, A. Palmer, L. Ständker, W.-G. Forssmann, J. Münch, J.C.M. Gebhardt, and L. Wiesmüller
Frontiers in Oncology 11, 689063 (2021), https://doi.org/10.3389/fonc.2021.689063
Altering transcription factor binding reveals comprehensive transcriptional kinetics of a basic gene
A.P. Popp, J. Hettich, and J.C.M. Gebhardt
Nucleic Acids Research 49, 6249–6266 (2021), https://doi.org/10.1093/nar/gkab443
Single molecule tracking and analysis framework including theory-predicted parameter settings
T. Kuhn*, J. Hettich*, R. Davtyan, and J.C.M. Gebhardt
Scientific Reports 11, 9465 (2021), (*equal contribution) https://doi.org/10.1038/s41598-021-88802-7
2020
Myosin VI moves on nuclear actin filaments and supports long-range chromatin rearrangements
A. Große-Berkenbusch, J. Hettich, T. Kuhn, N. Fili, A. Cook, Y. Hari-Gupta, A. Palmer, L. Streit, P. Ellis, C. Toseland#, and J.C.M. Gebhardt#
bioRxiv (2020), (#corresponding author) https://doi.org/10.1101/2020.04.03.023614
Inferring quantity and qualities of superimposed reaction rates from single molecule survival time distributions
M. Reisser*, J. Hettich*, T. Kuhn, A.P. Popp, A. Große-Berkenbusch, and J.C.M. Gebhardt
Scientific Reports 10, 1758 (2020), (*equal contribution) https://doi.org/10.1038/s41598-020-58634-y
2019
Mitotic chromosome binding predicts transcription factor properties in interphase
M. Raccaud, E.T. Friman, A.B. Alber, H. Agarwal, C. Deluz, T. Kuhn, J.C.M. Gebhardt, and D.M. Suter
Nature Communications 10, 487 (2019), https://doi.org/10.1038/s41467-019-08417-5
Single-molecule imaging of the transcription factor SRF reveals prolonged chromatin-binding kinetics upon cell stimulation
L. Hipp, J. Beer, O. Kuchler, M. Reisser, D. Sinske, J. Michaelis#, J.C.M. Gebhardt#, and B. Knöll#
PNAS 116, 880–889 (2019), (#corresponding author) https://doi.org/10.1073/pnas.1812734116
2018
Single-molecule imaging correlates decreasing nuclear volume with increasing TF-chromatin associations during zebrafish development
M. Reisser, A. Palmer, A.P. Popp, C. Jahn, G. Weidinger, and J.C.M. Gebhardt
Nature Communications 9, 5218 (2018), https://doi.org/10.1038/s41467-018-07731-8
Endonuclease G promotes mitochondrial genome cleavage and replication
R.S. Wiehe, B. Gole, L. Chatre, P. Walther, E. Calzia, A. Palmer, J.C.M. Gebhardt, M. Ricchetti, and L. Wiesmüller
Oncotarget 9, 89081 (2018), https://doi.org/10.18632/oncotarget.25645
Transcription factor target site search and gene regulation in a background of unspecific binding sites
J. Hettich and J.C.M. Gebhardt
Journal of Theoretical Biology 454, 91–101 (2018), https://doi.org/10.1016/j.jtbi.2018.05.037
2017
DNA residence time is a regulatory factor of transcription repression
K. Clauß*, A.P. Popp*, L. Schulze, J. Hettich, M. Reisser, L.E. Torres, N.H. Uhlenhaut, and J.C.M. Gebhardt
Nucleic Acids Research 45, 11121–11130 (2017), (*equal contribution) https://doi.org/10.1093/nar/gkx728
Direct Observation of Cell-Cycle-Dependent Interactions between CTCF and Chromatin
H. Agarwal, M. Reisser, C. Wortmann, and J.C.M. Gebhardt
Biophysical Journal 112, 2051–2055 (2017), https://doi.org/10.1016/j.bpj.2017.04.018
2014
Spatial organization of RNA polymerase II insidea mammalian cell nucleus revealed by reflectedlight-sheet superresolution microscopy
Z.W. Zhao*, R. Roy*, J.C.M. Gebhardt*, D.M. Suter*, A.R. Chapman, and X.S. Xie
PNAS 111, 681–686 (2014), (*equal contribution) https://doi.org/10.1073/pnas.1318496111
2013
From mechanical folding trajectories to intrinsic energy landscapes of biopolymers
M. Hinczewski, C.M. Gebhardt, M. Rief, and D. Thirumalai
PNAS 110, 4500–4505 (2013), https://doi.org/10.1073/pnas.1214051110
Reply to “convergence of chromatin binding estimates in live cells”
Z.W. Zhao, J.C.M. Gebhardt, D.M. Suter, and X.S. Xie
Nature Methods 10, 692 (2013), https://doi.org/10.1038/nmeth.2574
Single-molecule imaging of transcription factor binding to DNA in live mammalian cells
J.C.M. Gebhardt*, D.M. Suter*, R. Roy, Z.W. Zhao, A.R. Chapman, S. Basu, T. Maniatis, and X.S. Xie
Nature Methods 10, 421–426 (2013), (*equal contribution) https://doi.org/10.1038/nmeth.2411
2011
The complex folding network of single calmodulin molecules
J. Stigler, F. Ziegler, A. Gieseke, J.C.M. Gebhardt, and M. Rief
Science 334, 512–516 (2011), https://doi.org/10.1126/science.1207598
2010
Regulation of a heterodimeric kinesin-2 through an unprocessive motor domain that is turned processive by its partner
M. Brunnbauer, F. Mueller-Planitz, S. Kösem, T.H. Ho, R. Dombi, J.C.M. Gebhardt, M. Rief, and Z. Ökten
PNAS 107, 10460–10465 (2010), https://doi.org/10.1073/pnas.1005177107
Full distance-resolved folding energy landscape of one single protein molecule
J.C.M. Gebhardt, T. Bornschlögl, and M. Rief
PNAS 107, 2013–2018 (2010), https://doi.org/10.1073/pnas.0909854107
The lever arm effects a mechanical asymmetry of the myosin-V-actin bond
J.C.M. Gebhardt, Z. Ökten, and M. Rief
Biophysical Journal 98, 277–281 (2010), https://doi.org/10.1016/j.bpj.2009.10.017
2009
Designing the folding mechanics of coiled coils
T. Bornschlögl, J. Christof M Gebhardt, and M. Rief
ChemPhysChem 10, 2800–2804 (2009), https://doi.org/10.1002/cphc.200900575
Force signaling in biology
J.C.M. Gebhardt, and M. Rief
Science 324, 1278–1280 (2009), https://doi.org/10.1126/science.1175874
2007
Myosin V stepping mechanism
G. Cappello, P. Pierobon, C. Symonds, L. Busoni, J.C.M. Gebhardt, M. Rief, and J. Prost
PNAS 104, 15328–15333 (2007), https://doi.org/10.1073/pnas.0706653104
2006
Myosin-V is a mechanical ratchet
J.C.M. Gebhardt, A.E.M. Clemen, J. Jaud, and M. Rief
PNAS 103, 8680–8685 (2006), https://doi.org/10.1073/pnas.0510191103
2003
The smallest possible nanocrystals of semiionic oxides
P. Persson, J.C.M. Gebhardt, and S. Lunell
Journal of Physical Chemistry B 107, 3336–3339 (2003), https://doi.org/10.1021/jp022036e
| Professors: | Prof. Dr. J. Christof M. Gebhardt Prof. Dr. Kay-Eberhard Gottschalk | ||||
| Senior-Professors: | Prof. Dr. sc. nat./ETH Zürich Othmar Marti Prof. Dr. Heinrich Hörber | ||||
| Secretariat: | N25 / 5208
Tel.: +49 (0)731 50 23010 | ||||
| Address: | Institute of Experimental Physics Albert-Einstein-Allee 11 89081 Ulm |
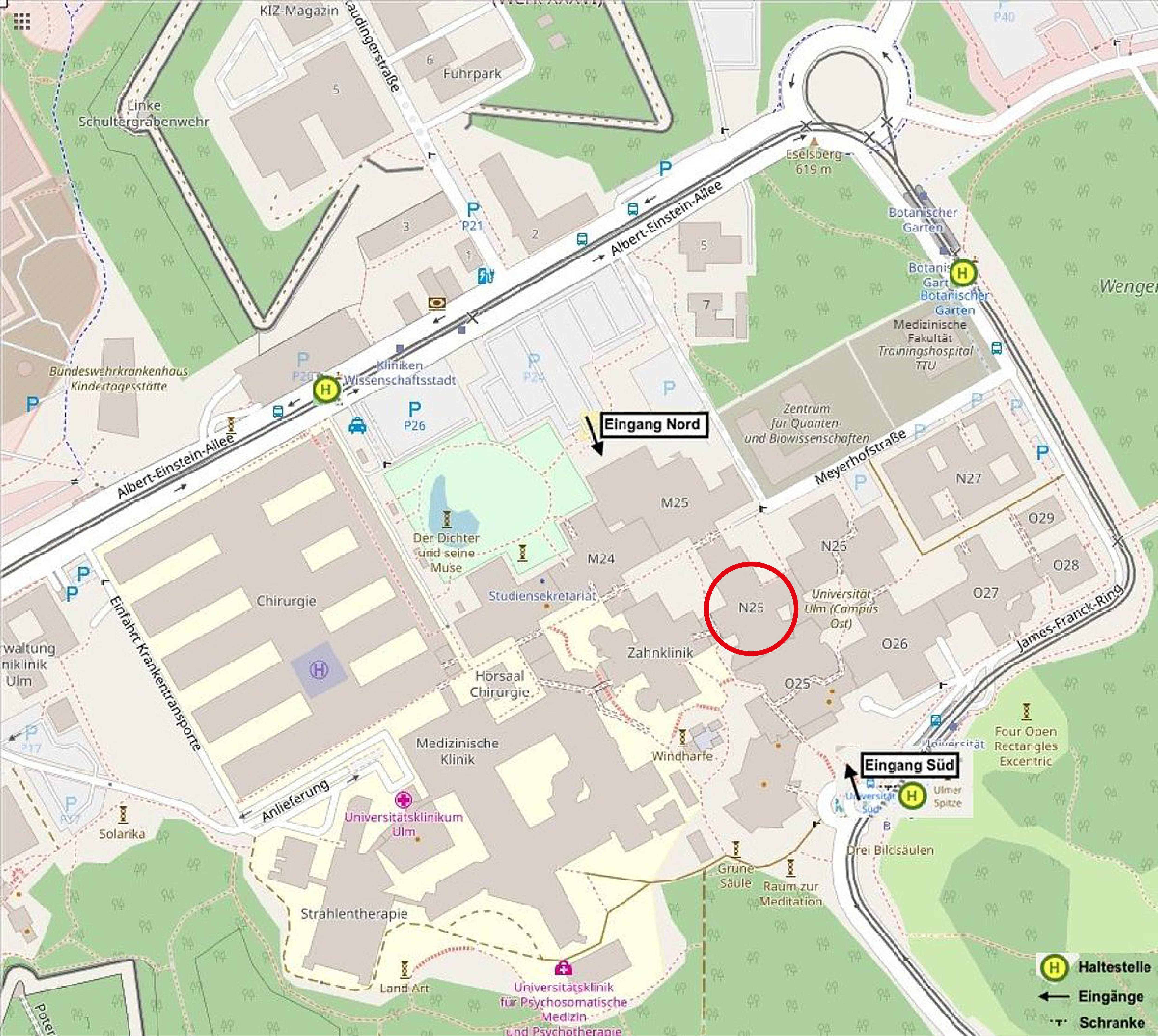
How to reach Ulm University:
Directions, Campusnavigator
e.g. with the tram:
From Ulm main station, every 10 minutes, take tram 2 towards Science Park II until stop Universität Süd.
How to find the institute:
The Institute of Experimental Physics is located in building part N25 of the university East on level 5. The secretariat is in room N25/5208.
from South entrance: go through the entire entry hall towards the North until the staircase/lift of N25.
from North entrance: go left (East) to M25, then right (South) to the staircase/lift of N25.
from East entrance: go left (West) to the staircase/lift of N25.
If you are lost: call the secretariat via 0731 50 23010.
