Contact
Dr. Medhanie Mulaw
Tel: 0731 502 6728
The advent of single-cell sequencing has further enhanced the importance of NGS and has enabled investigators to ask questions that would normally not be feasible to address via bulk sequencing. Single-cell sequencing methodologies enable the analysis of the transcriptome, mutatome, protein–DNA interaction, and broadly the epigenome. Combined with increased statistical power and advanced analytic tools geared toward single-cell analysis, one can then look at, but not limited to, tissue heterogeneity, clonality, analysis of gene- and allele-specific expression, and single-cell level mutational analysis. More recently, combinatorial approaches have been developed that would allow simultaneous interrogation of macromolecules (multi-omics) and spatial context of cells. This is especially highly interesting in tissues and systems with relatively higher complexity and heterogeneity.
The single-cell sequencing unit currently provides bioinformatics analysis services for single-cell datasets generated using current state-of-the-art technologies.
Services
1. Uni-modal single-cell (scRNA-seq, scATAC-seq, sc immunome, spatial transcriptomics …)
Project-specific analysis is performed. Some of the analysis methods used include:
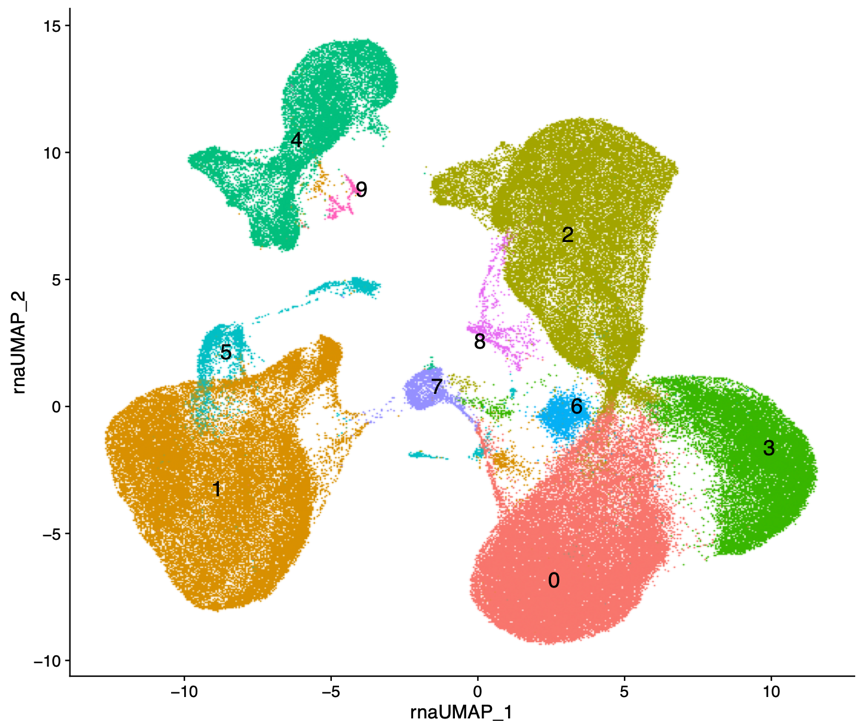
(a) Identification of clusters and neighbors (PCA and UMAPs), marker identification and cell annotations, trajectory and velocity analysis, scGSEA, cell-cell communication, pseudobulk, and integrative analysis with in-house or published bulk sequencing datasets.
scATAC-seq specific analyses: Differential accessibility analysis, classification of differentially accessible regions by genomics regions, annotation, and motif enrichment analysis, comparative and integrative analysis with related scRNA or bulk RNA-seq datasets.
Spatial transcriptomics specific analyses: Identification of Variable Features by regions, Integration with single-cell data reference including Spatial deconvolution.
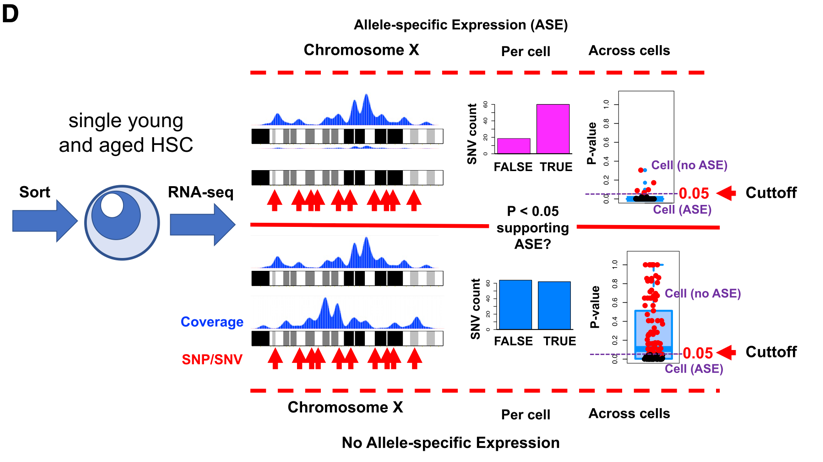
(b) Allele-specific expression analysis: using discriminant SNPs as markers, we quantify allele-specific expression of genes in scRNA-seq datasets (see Fig. 2 and our study Gregorian et al. (2021)).
i. The number of reads covering high-confidence heterozygous single nucleotide variants (SNVs) according to a selected organismal database
ii. Positional (chromosomal) enrichment analysis of genes with significant ASE evidence
iii. Functional enrichment analysis and association analysis with relevant sequencing datasets reflecting genome organization (e.g. ATAC-seq)
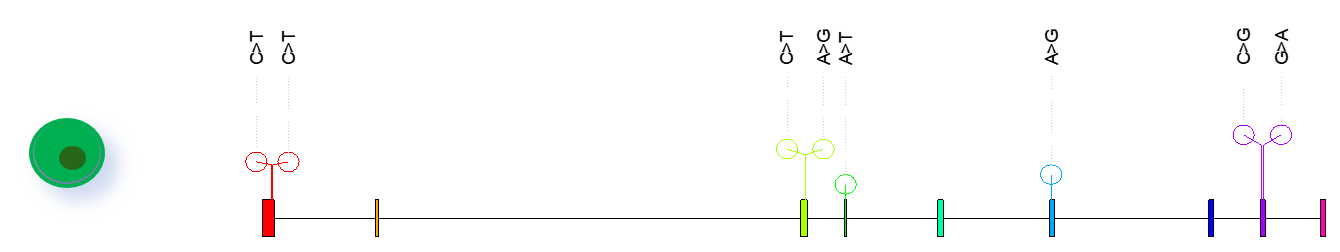
(c) Single-cell mutational and clonality analysis: We have developed efficient single-cell mutational analysis approaches combined with clonality and somatic mutational signature analysis (Florian et al. 2016). We can also provide expressed variants analysis for plate-based full transcript single-cell RNA-seq data (see Fig. 3).
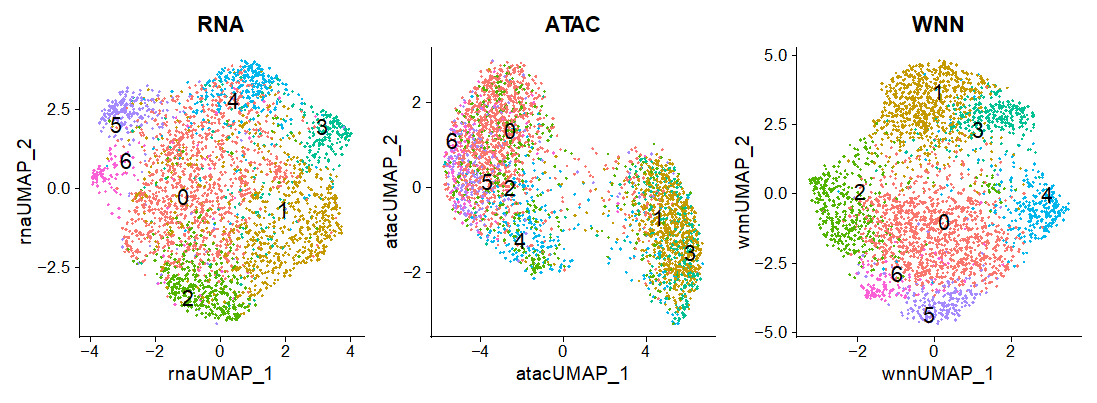
2. Multimodal single-cell approaches (RNA+ATAC/share-seq, CITE-seq …)
Integrated analysis (Multimodal neighbors) of multimodal data followed by analysis steps discussed under 1. Furthermore, multimodal specific analysis including promoter accessibility and gene expression analysis (share-seq), surface marker, and transcriptome integrated analysis (CITE-seq).
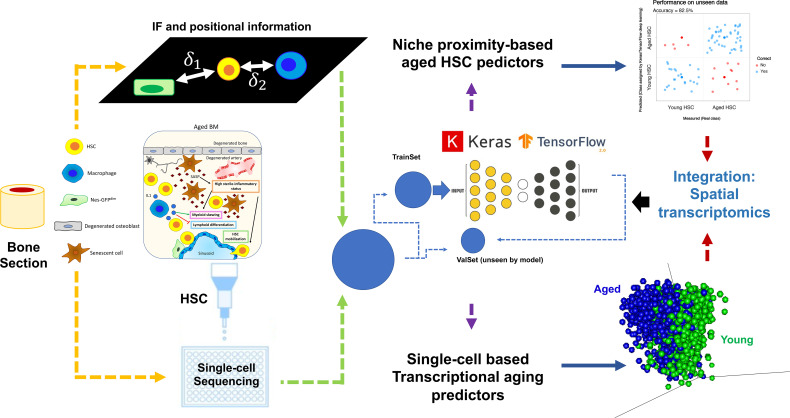
3. Machine learning (ML) and AI methodologies
Machine learning (ML) and AI methodologies in single-cell genomics including integrative analysis with in-vitro, in-vivo, and/or clinical datasets.
We provide ML/AI approaches suitable to single-cell omics (classification/prediction/marker identification). Such data can also be integrated with in-vitro, in-vivo, and/or clinical datasets to identify markers defining molecular mechanisms and clinical readouts (see Fig. 5 and our review Matteini, Mulaw, and Florian, 2021).