Research Topics
Identification and characterization of novel AMPs/bacteriocins
Listeria monocytogenes is a ubiquitous, Gram-positive soil bacterium and a frequent contaminant of processed food products. Its abilities to withstand and grow under a wide range of environmental conditions (stress resistance to high/low temperature, pH, salt, etc.) and to form biofilms on different surfaces make L. monocytogenes a serious concern in the food processing industries. Moreover, L. monocytogenes is able to cause life-threatening infections in immuno-compromised persons and serves as a widely used model organism for intracellular pathogenesis.
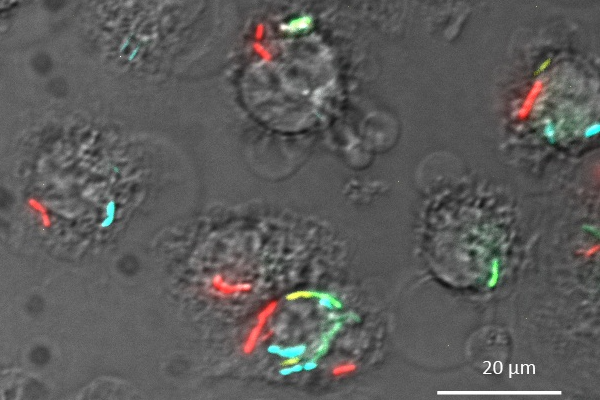
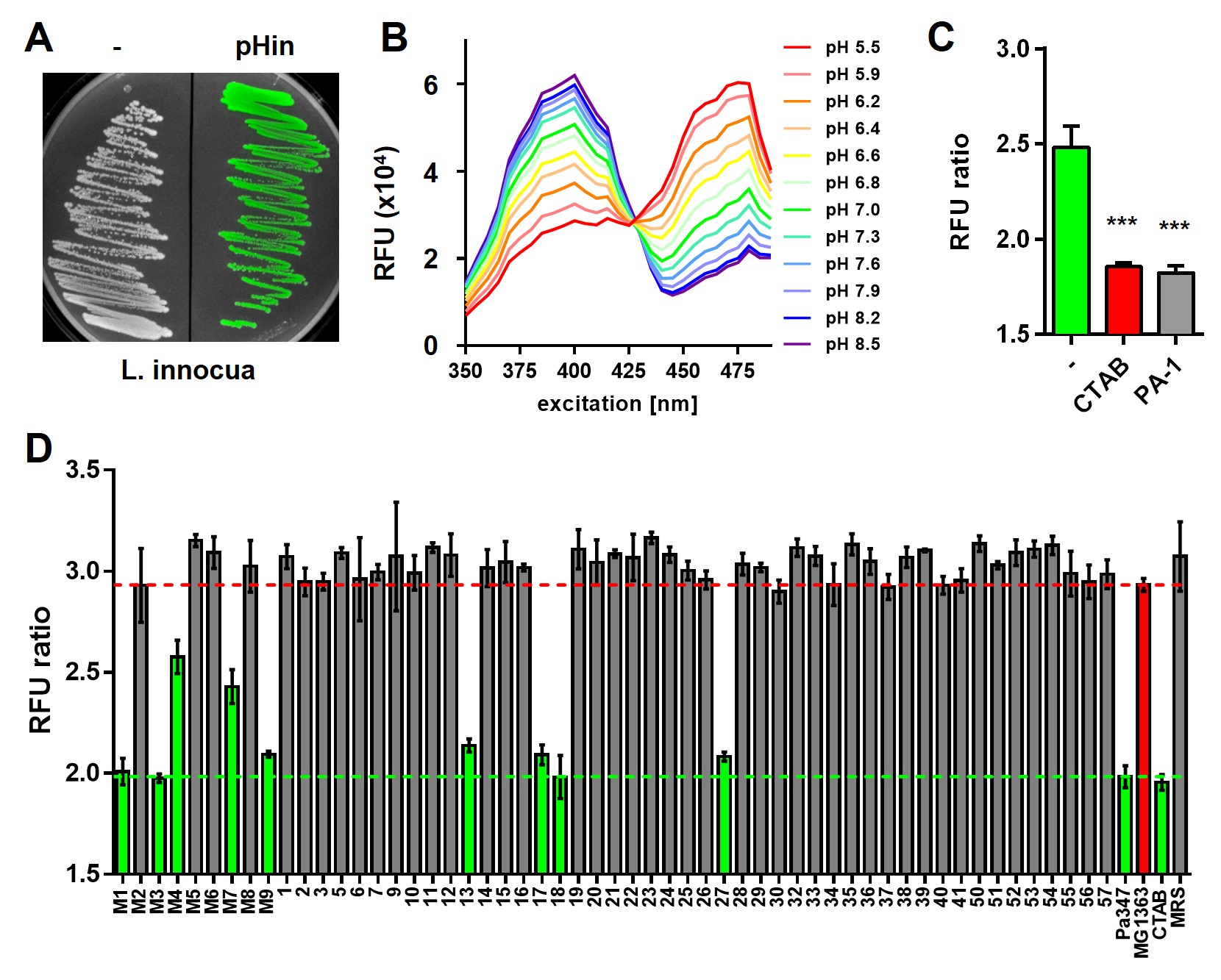
In recent years, our group was involved in a European research consortium that has investigated the stress response of L. monocytogenes to high pressure processing (HPP) used for food preservation. This allowed identification of a number of genes as potential targets for pretreatment targets to improve the efficiency of high-pressure treatment (Duru et al., 2021a, 2021b, 2020; Nikparvar et al., 2021a, 2021b). Interestingly, one group of genes differentially regulated by HPP are mannose-specific phosphotransferase systems, which are known as receptors for AMPs belonging to class IIa bacteriocins of the pediocin family. As part of this research consortium, we have constructed sensor strains of human pathogens such as L. monocytogenes (Crauwels et al., 2018) and related non-pathogenic model organisms (e.g. Listeria innocua) that allow to monitoring of intracellular pH (Figure 1). The measurements are based on expression of fluorescent proteins that show changes in the fluorescent properties in a pH-dependent manner. Thus, rapid changes of the intracellular (neutral) pH of the sensor bacteria to the external pH in a more acidic or alkaline environment/buffer are indicative of a disruption of membrane integrity. For example, these sensor strains can be used to screen supernatants of bacterial strain collections for AMPs with pore-forming activity as recently demonstrated for a small library of raw milk isolates (Figure 1 and Desiderato et al., 2021).
In different projects involving both academic and industrial partners, these sensor bacteria are used to screen larger collections of bacteria, e.g. starter cultures for food production, to identify producers of (novel) bacteriocins. Alternatively, gene clusters for biosynthesis of bacteriocins and corresponding resistance mechanisms can be identified in genome and metagenome sequences databases using predictive software tools (Goldbeck et al., 2021b). Once identified, the bacteriocins produced by these bacteria, their mechanisms of action and receptors can be analyzed and characterized again using the developed sensor bacteria.
Biotechnological production of bacteriocins
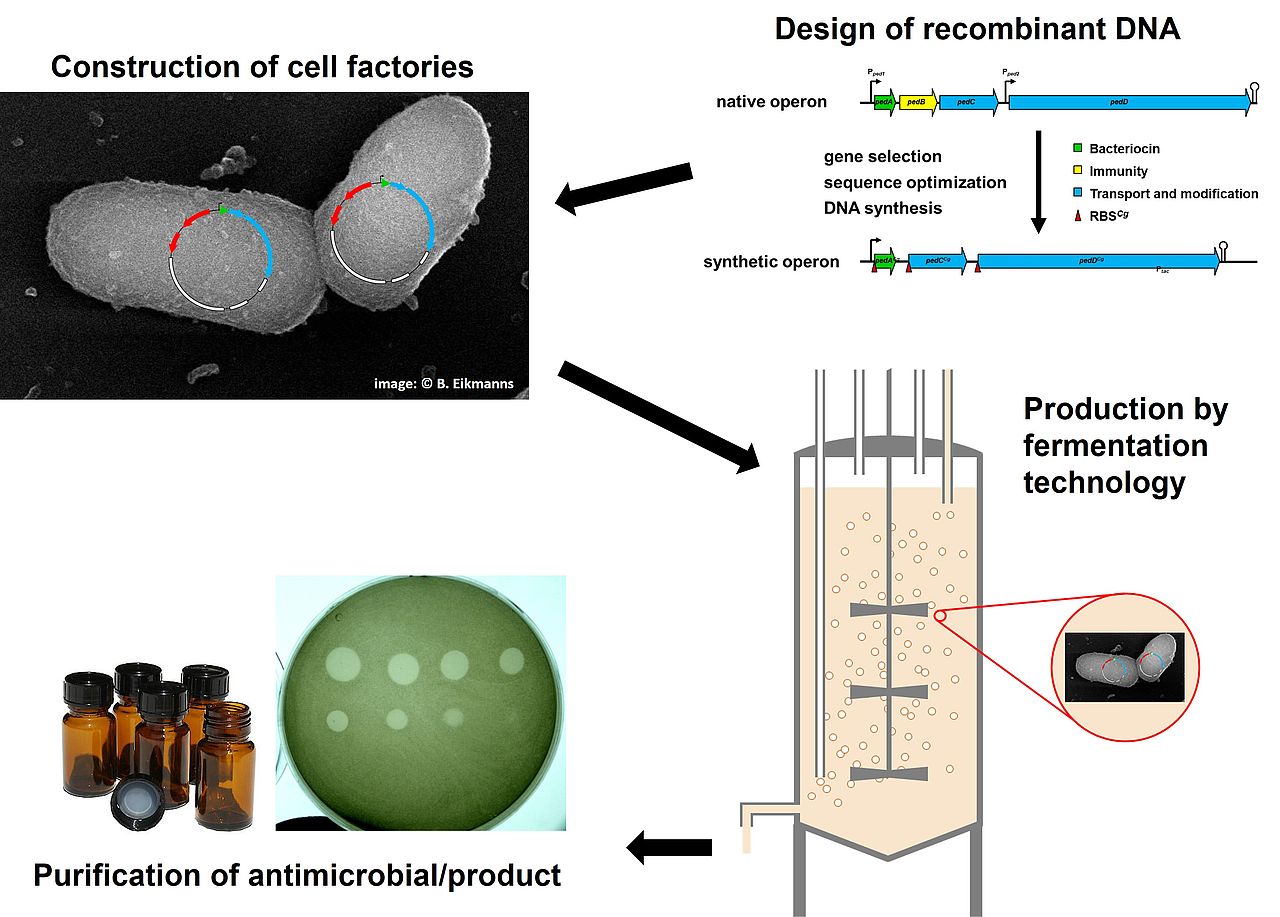
We are also investigating strategies to develop the findings of our basic research into health-related products. Bacteriocins are antimicrobial peptides (AMPs) produced by bacteria to inhibit competitors in their natural environments. Several bacteriocins are widely used as food preservatives. Due to the rapid increase in antibiotic resistant bacteria, bacteriocins are also discussed as alternatives to antibiotics to treat infections for therapeutic purposes. Currently, industrial production of bacteriocins is performed exclusively with natural producer organisms on complex substrates and products are commercialized as semi-purified preparations or crude fermentates. To allow clinical application and entry of novel bacteriocins into the market, efficacy of production and purity of the product need to be improved. One possibility is to shift production to recombinant biotechnological production host. Due to the intrinsic antimicrobial activity of bacteriocins, this is not a trivial task. Recently, were able to establish recombinant production of bacteriocins using the widely used industrial workhorse organisms Corynebacterium glutamicum as host (Goldbeck et al., 2021a). Pediocin PA-1 is a linear class IIa bacteriocin without extensive modifications produced naturally by Pediococcus spp. and Lactobacillus spp. strains. An even more interesting bacteriocin from a scientific and commercial perspective is nisin, which is the most intensively studied bacteriocin. Nisin is widely used as a food preservative and is the bacteriocin with the largest market volume worldwide. It has broad antimicrobial activity against several human pathogens. However, nisin is more difficult to produce using recombinant hosts due to its high degree of posttranslational modification and, unfortunately, it is also active against C. glutamicum. In order to address these obstacles we study resistance mechanisms of bacteria against nisin and other lantibiotics (Goldbeck et al., 2021b) aiming to transfer these resistance mechanisms to C. glutamicum (Weixler et al., 2021). Additionally, we are looking for alternative approaches such as production of inactive precursors and downstream activation.
Molecular tools for bifidobacteria to study their effects on human health
Bifidobacteria are important group of anaerobic, Gram-positive bacteria of the lower intestinal tract of humans particularly abundant in breast-fed infants. Various health-promoting properties are attributed to the presence of bifidobacteria in the gut. Based on these health-promoting properties, bifidobacteria are widely used as so-called probiotic supplements in pharmaceutical and dairy products. For some strains, anti-inflammatory effects have been reported in vitro and in preclinical studies. Thus, one potential field of application is their use as alternative or supplementary treatment in inflammatory conditions of the gastrointestinal tract.
In previous studies, our group has identified Bifidobacterium spp. strains that show high adhesion to intestinal epithelial cells and display potent anti-inflammatory activity in vitro and in murine models of colitis (Grimm et al., 2015; Philippe et al., 2011; Preising et al., 2010). To unravel the molecular mechanisms of host colonization and anti-inflammatory activity, we sequenced the genomes of several strains of bifidobacteria (Bottacini et al., 2010; Zhurina et al., 2013, 2011) and identified a number of candidate genes potentially involved in these properties (Westermann et al., 2012). Past and current projects aim at investigating the role of these candidates to adhesion, colonization and inhibition of inflammation (Gleinser et al., 2012; Grimm et al., 2015; Wei et al., 2014). A further aspect is the potential use of bifidobacteria as vectors to for delivery of therapeutic genes into solid tumors (Osswald et al., 2015a).
Due to the notorious resistance of bifidobacteria to genetic manipulation, especially the generation of defined mutant strains, we are mainly active in developing new molecular tools for these organisms (Figure 3; Grimm et al., 2014; Osswald et al., 2015b; Sun et al., 2014). Currently we are exploring the possibility for precise genetic engineering of bifidobacteria e.g. using CRISPR/Cas-based molecular tools.