Publications 2018
Single-molecule imaging of the transcription factor SRF reveals prolonged chromatin-binding kinetics upon cell stimulation
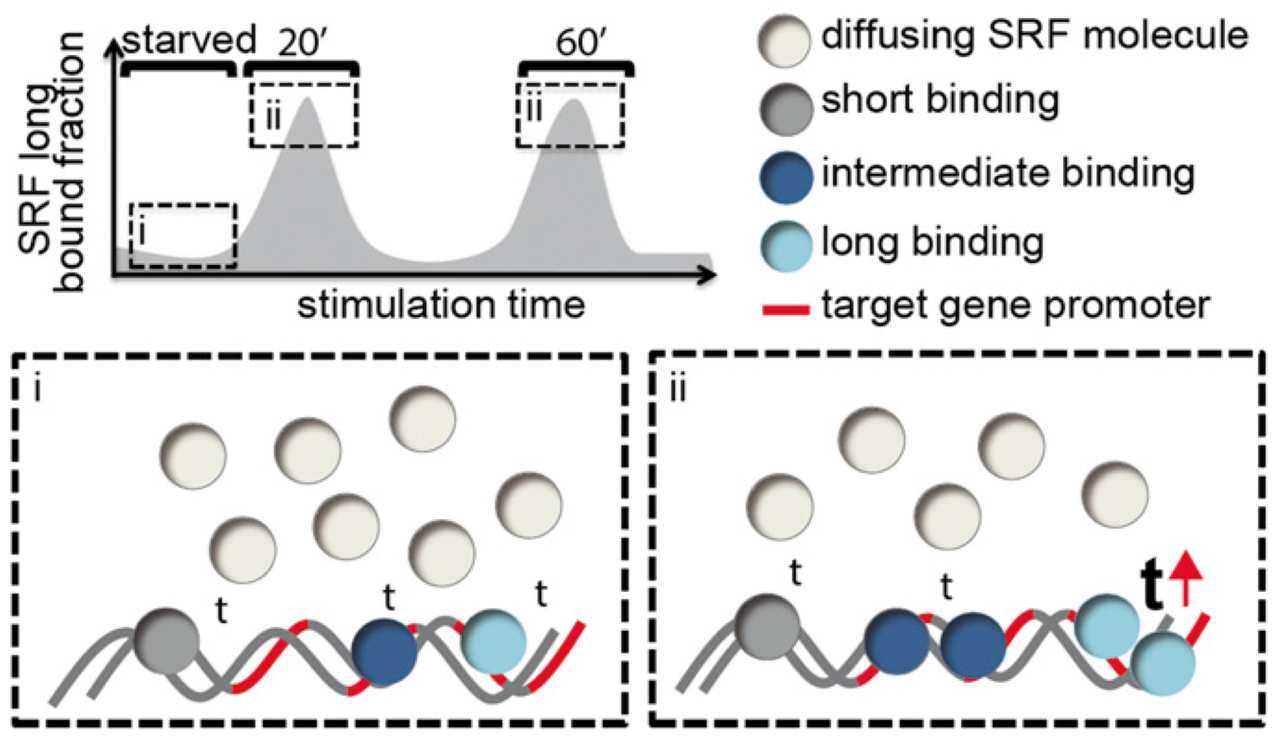
Lisa Hipp, Judith Beer, Oliver Kuchler, Matthias Reisser, Daniela Sinske, Jens Michaelis, J. Christof M. Gebhardt, and Bernd Knöll
Serum response factor (SRF) mediates immediate early gene (IEG) and cytoskeletal gene expression programs in almost any cell type. So far, SRF transcriptional dynamics have not been investigated at single-molecule resolution. We provide a study of single Halo-tagged SRF molecules in fibroblasts and primary neurons. In both cell types, individual binding events of SRF molecules segregated into three chromatin residence time regimes, short, intermediate, and long binding, indicating a cell type-independent SRF property. The chromatin residence time of the long bound fraction was up to 1 min in quiescent cells and significantly increased upon stimulation. Stimulation also enhanced the long bound SRF fraction at specific timepoints (20 and 60 min) in both cell types. These peaks correlated with activation of the SRF cofactors MRTF-A and MRTF-B (myocardin-related transcription factors). Interference with signaling pathways and cofactors demonstrated modulation of SRF chromatin occupancy by actin signaling, MAP kinases, and MRTFs.
PNAS
doi.org/10.1073/pnas.1812734116
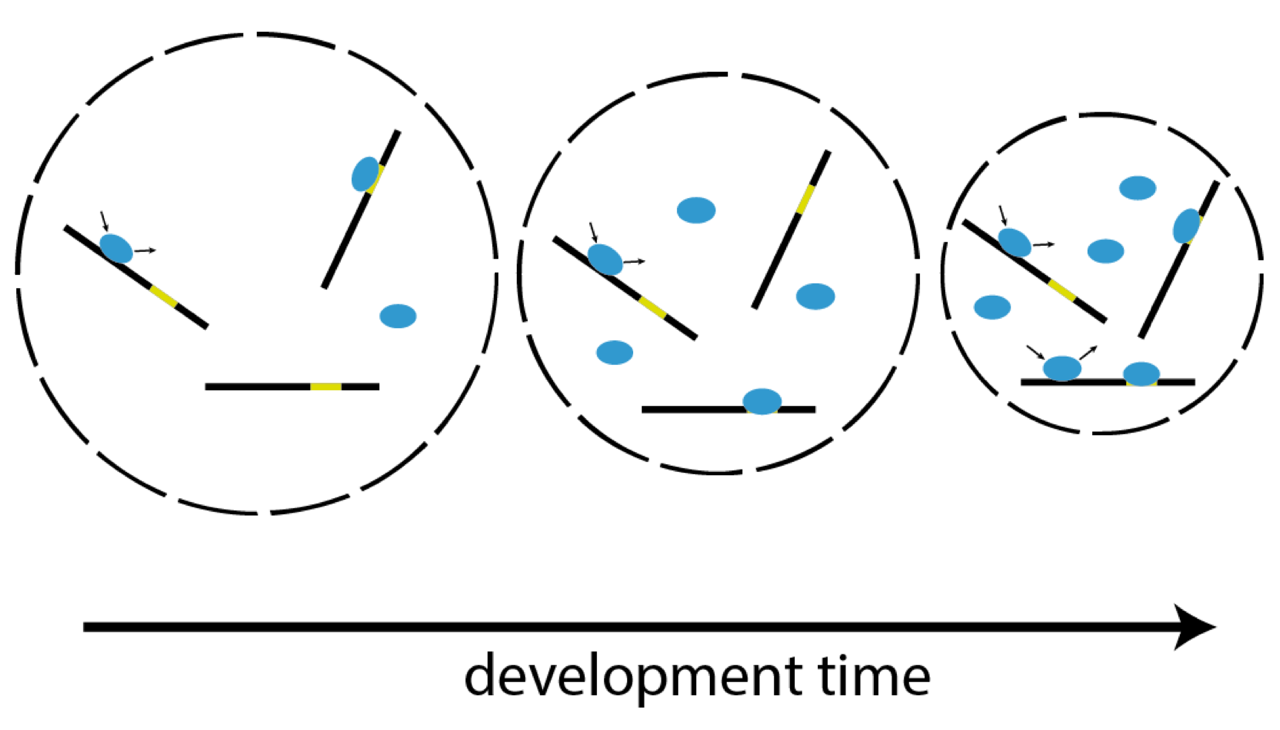
Single-molecule imaging correlates decreasing nuclear volume with increasing TF-chromatin associations during zebrafish development
Matthias Reisser, Anja Palmer, Achim P. Popp, Christopher Jahn, Gilbert Weidinger & J. Christof M. Gebhardt
Zygotic genome activation (ZGA), the onset of transcription after initial quiescence, is a major developmental step in many species, which occurs after ten cell divisions in zebrafish embryos. How transcription factor (TF)-chromatin interactions evolve during early development to support ZGA is largely unknown. We establish single molecule tracking in live developing zebrafish embryos using reflected light-sheet microscopy to visualize two fluorescently labeled TF species, mEos2-TBP and mEos2-Sox19b. We further develop a data acquisition and analysis scheme to extract quantitative information on binding kinetics and bound fractions during fast cell cycles. The chromatin-bound fraction of both TFs increases during early development, as expected from a physical model of TF-chromatin interactions including a decreasing nuclear volume and increasing DNA accessibility. For Sox19b, data suggests the increase is mainly due to the shrinking nucleus. Our single molecule approach provides quantitative insight into changes of TF-chromatin associations during the developmental period embracing ZGA.
Nature Communications
www.nature.com/articles/s41467-018-07731-8
Complete Kinetic Theory of FRET

Tobias Eilert, Eleni Kallis, Julia Nagy, Carlheinz Röcker and Jens Michaelis
Förster resonance energy transfer (FRET) can be used to measure distances and infer structures at the molecular level. However, the flexible linkers with which the fluorophores are attached to a macromolecule introduce a lack of knowledge. Both the dye’s geometry and kinetics give rise to uncertainties. Whereas the impact of the geometry is already well understood, the real extent of the kinetics has not been investigated thoroughly. Here, we present a single-molecule (sm)FRET theory that defines the kinetics of dye movements in a complete form. We introduce a formal nomenclature and provide a recipe for the calculation of the corresponding FRET efficiency. We further analyze experimental data in order to obtain parameters characterizing the geometry and kinetics of the FRET dyes and use them to resimulate the FRET efficiencies by diffusion of fluorophore and linker movement. We show in a real case scenario of dye molecules attached to dsDNA that when making geometrical and kinetic assumptions commonly used in the FRET community one obtains results differing from the experimental data. In contrast, our stochastic simulations taking kinetic parameters from experiments into account reproduce the correct FRET efficiencies. Furthermore, we present a method enabling us to classify the kinetics of the dyes by investigating single realizations of the simulated transfer process. The results support our notion that the common kinetic assumptions are not appropriate over the whole range of distances inferred by FRET even for the simple situation of dyes attached to DNA where few interactions occur.
J. Phys. Chem. B
https://pubs.acs.org/articlesonrequest/AOR-PGPEUJMdg5vms738vgRS
Precision and accuracy of single-molecule FRET measurements—a multi-laboratory benchmark study
Björn Hellenkamp, Sonja Schmid, Olga Doroshenko, Oleg Opanasyuk, Ralf Kühnemuth, Soheila Rezaei Adariani, Benjamin Ambrose, Mikayel Aznauryan, Anders Barth, Victoria Birkedal, Mark E. Bowen, Hongtao Chen, Thorben Cordes, Tobias Eilert, Carel Fijen, Christian Gebhardt, Markus Götz, Giorgos Gouridis, Enrico Gratton, Taekjip Ha, Pengyu Hao, Christian A. Hanke, Andreas Hartmann, Jelle Hendrix, Lasse L. Hildebrandt, Verena Hirschfeld, Johannes Hohlbein, Boyang Hua, Christian G. Hübner, Eleni Kallis, Achillefs N. Kapanidis, Jae-Yeol Kim, Georg Krainer, Don C. Lamb, Nam Ki Lee, Edward A. Lemke, Brié Levesque, Marcia Levitus, James J. McCann, Nikolaus Naredi-Rainer, Daniel Nettels, Thuy Ngo, Ruoyi Qiu, Nicole C. Robb, Carlheinz Röcker, Hugo Sanabria, Michael Schlierf, Tim Schröder, Benjamin Schuler, Henning Seidel, Lisa Streit, Johann Thurn, Philip Tinnefeld, Swati Tyagi, Niels Vandenberk, Andrés Manuel Vera, Keith R. Weninger, Bettina Wünsch, Inna S. Yanez-Orozco, Jens Michaelis, Claus A. M. Seidel, Timothy D. Craggs & Thorsten Hugel
Single-molecule Förster resonance energy transfer (smFRET) is increasingly being used to determine distances, structures, and dynamics of biomolecules in vitro and in vivo. However, generalized protocols and FRET standards to ensure the reproducibility and accuracy of measurements of FRET efficiencies are currently lacking. Here we report the results of a comparative blind study in which 20 labs determined the FRET efficiencies (E) of several dye-labeled DNA duplexes. Using a unified, straightforward method, we obtained FRET efficiencies with s.d. between ±0.02 and ±0.05. We suggest experimental and computational procedures for converting FRET efficiencies into accurate distances, and discuss potential uncertainties in the experiment and the modeling. Our quantitative assessment of the reproducibility of intensity-based smFRET measurements and a unified correction procedure represents an important step toward the validation of distance networks, with the ultimate aim of achieving reliable structural models of biomolecular systems by smFRET-based hybrid methods.
Nature Methods
www.nature.com/articles/s41592-018-0085-0

The blue and red spotlights represent the twenty-seven institutions around the world, which blindly measured distances within DNAs with Angstrom precision. One of the blind samples wraps around the globe to bring together countries and time-zones as an example of what science without borders can accomplish. The dyes are shown as diffuse clFigure: The blue and red spotlights represent the twenty-seven institutions around the world, which blindly measured distances within DNAs with Angstrom precision. One of the blind samples wraps around the globe to bring together countries and time-zones as an example of what science without borders can accomplish. The dyes are shown as diffuse clFigure: The blue and red spotlights represent the twenty-seven institutions around the world, which blindly measured distances within DNAs with Angstrom precision. One of the blind samples wraps around the globe to bring together countries and time-zones as an example of what science without borders can accomplish. The dyes are shown as diffuse clouds over the surface of the DNA. Designed by Hugo Sanabria and Nandakumar Chedikulathu Vishnu (Clemson University).
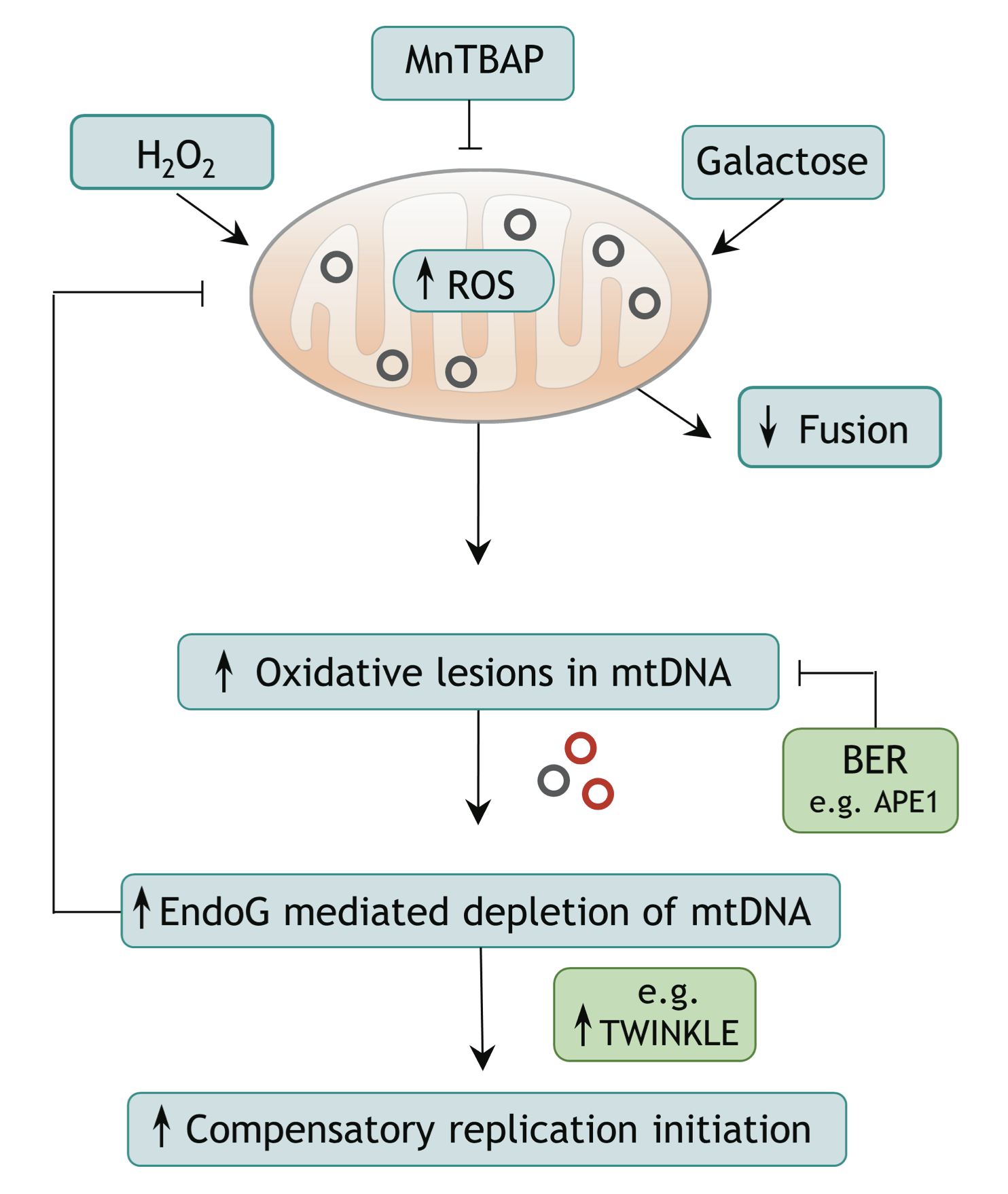
Endonuclease G promotes mitochondrial genome cleavage and replication
Rahel Stefanie Wiehe, Boris Gole, Laurent Chatre, Paul Walther, Enrico Calzia, Anja Palmer, J. Christof M. Gebhardt, Miria Ricchetti and Lisa Wiesmüller
Endonuclease G (EndoG) is a nuclear-encoded endonuclease, mostly localised in mitochondria. In the nucleus EndoG participates in site-specific cleavage during replication stress and genome-wide DNA degradation during apoptosis. However, the impact of EndoG on mitochondrial DNA (mtDNA) metabolism is poorly understood. Here, we investigated whether EndoG is involved in the regulation of mtDNA replication and removal of aberrant copies. We applied the single-cell mitochondrial Transcription and Replication Imaging Protocol (mTRIP) and PCR-based strategies on human cells after knockdown/knockout and re-expression of EndoG. Our analysis revealed that EndoG stimulates both mtDNA replication initiation and mtDNA depletion, the two events being interlinked and dependent on EndoG’s nuclease activity. Stimulation of mtDNA replication by EndoG was independent of 7S DNA processing at the replication origin. Importantly, both mtDNA-directed activities of EndoG were promoted by oxidative stress. Inhibition of base excision repair (BER) that repairs oxidative stress- induced DNA damage unveiled a pronounced effect of EndoG on mtDNA removal, reminiscent of recently discovered links between EndoG and BER in the nucleus. Altogether with the downstream effects on mitochondrial transcription, protein expression, redox status and morphology, this study demonstrates that removal of damaged mtDNA by EndoG and compensatory replication play a critical role in mitochondria homeostasis.
Oncotarget
doi.org/10.18632/oncotarget.24822
Dysregulation of a novel miR-1825/TBCB/TUBA4A pathway in sporadic and familial ALS.
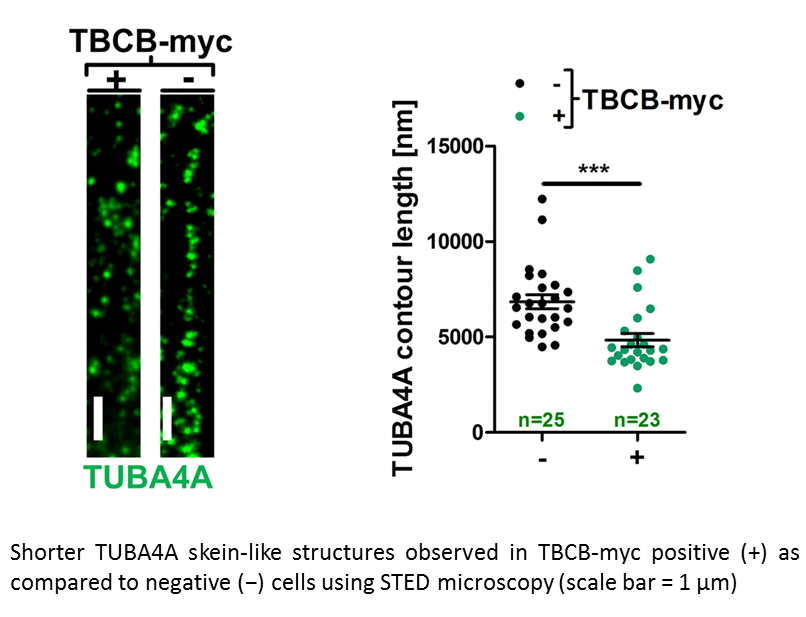
Anika M. Helferich, Sarah J. Brockmann, Jörg Reinders, Dhruva Deshpande, Karlheinz Holzmann, David Brenner, Peter M. Andersen, Susanne Petri, Dietmar R. Thal, Jens Michaelis, Markus Otto, Steffen Just, Albert C. Ludolph, Karin M. Danzer, Axel Freischmidt, Jochen H. Weishaupt
Genetic and functional studies suggest diverse pathways being affected in the neurodegenerative disease amyotrophic lateral sclerosis (ALS), while knowledge about converging disease mechanisms is rare. We detected a downregulation of microRNA-1825 in CNS and extra-CNS system organs of both sporadic (sALS) and familial ALS (fALS) patients. Combined transcriptomic and proteomic analysis revealed that reduced levels of microRNA-1825 caused a translational upregulation of tubulin-folding cofactor b (TBCB). Moreover, we found that excess TBCB led to depolymerization and degradation of tubulin alpha-4A (TUBA4A), which is encoded by a known ALS gene. Importantly, the increase in TBCB and reduction of TUBA4A protein was confirmed in brain cortex tissue of fALS and sALS patients, and led to motor axon defects in an in vivo model. Our discovery of a microRNA-1825/TBCB/TUBA4A pathway reveals a putative pathogenic cascade in both fALS and sALS extending the relevance of TUBA4A to a large proportion of ALS cases.
Cellular and Molecular Life Sciences
doi.org/10.1007/s00018-018-2873-1
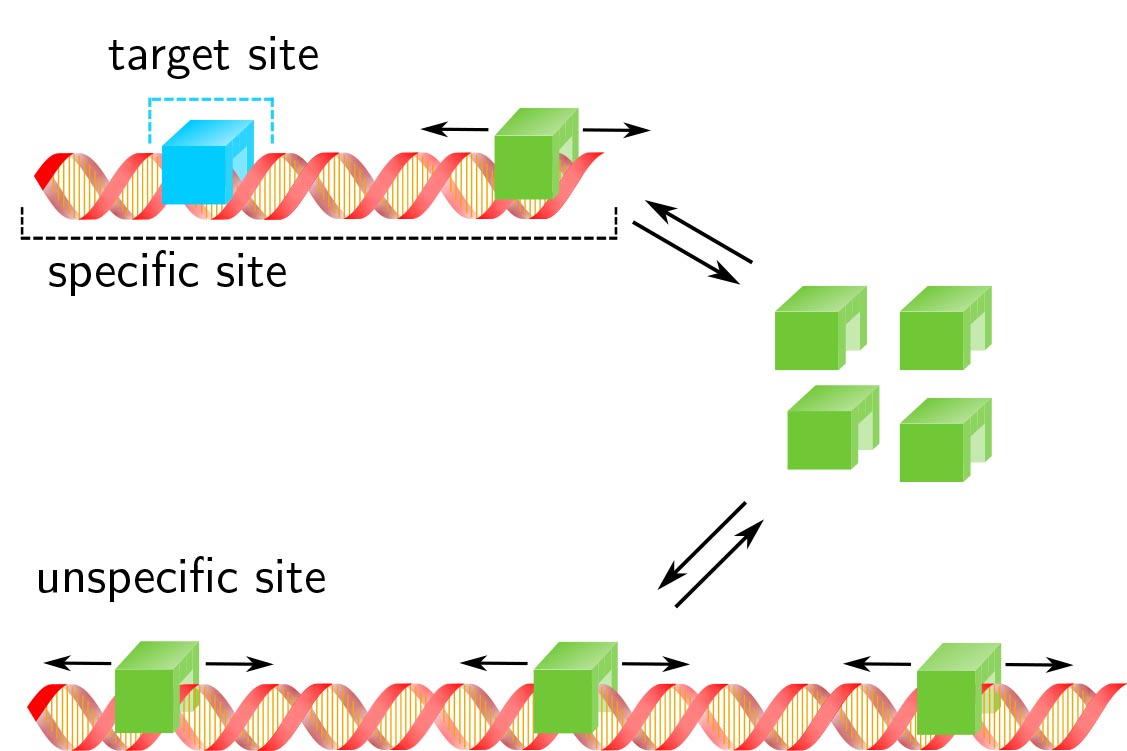
Transcription factor target site search and gene regulation in a background of unspecific binding sites
J. Hettich J.C.M. Gebhardt
Response time and transcription level are vital parameters of gene regulation. They depend on how fast transcription factors (TFs) find and how efficient they occupy their specific target sites. It is well known that target site search is accelerated by TF binding to and sliding along unspecific DNA and that unspecific associations alter the occupation frequency of a gene. However, whether target site search time and occupation frequency can be optimized simultaneously is mostly unclear. We developed a transparent and intuitively accessible state-based formalism to calculate search times to target sites on and occupation frequencies of promoters of arbitrary state structure. Our formalism is based on dissociation rate constants experimentally accessible in live cell experiments. To demonstrate our approach, we consider promoters activated by a single TF, by two coactivators or in the presence of a competitive inhibitor. We find that target site search time and promoter occupancy differentially vary with the unspecific dissociation rate constant. Both parameters can be harmonized by adjusting the specific dissociation rate constant of the TF. However, while measured DNA residence times of various eukaryotic TFs correspond to a fast search time, the occupation frequencies of target sites are generally low. Cells might tolerate low target site occupancies as they enable timely gene regulation in response to a changing environment.
Journal of Theoretical Biology
doi.org/10.1016/j.jtbi.2018.05.037
Chemoselective Dual Labeling of Native and Recombinant Proteins
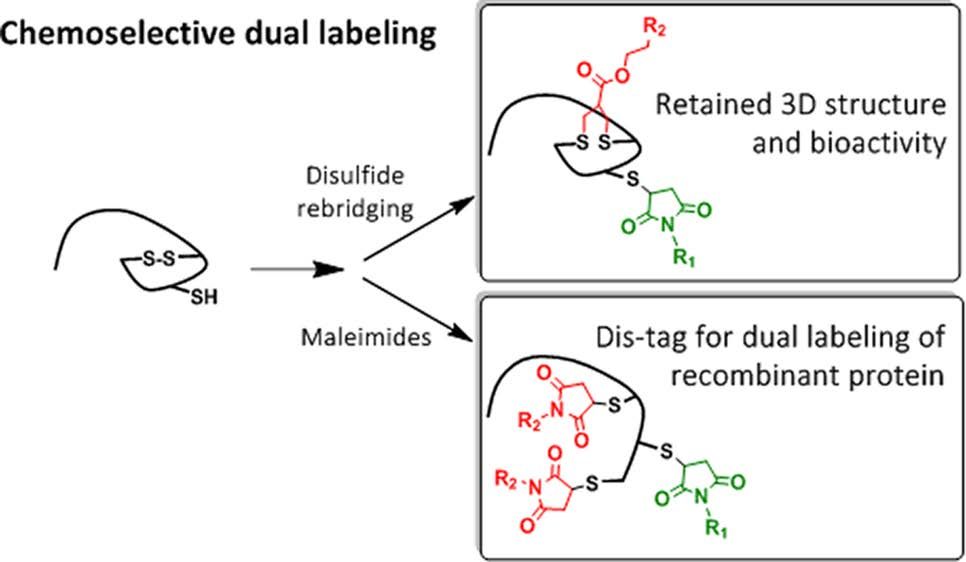
Bikram Keshari Agrawalla, Tao Wang, Andreas Riegger, Matthias P. Domogalla, Kerstin Steinbrink, Thilo Dörfler, Xi Chen†, Felix Bold, Markus Lamla, Jens Michaelis, Seah Ling Kuan, Tanja Weil
The attachment of two different functionalities in a site-selective fashion represents a great challenge in protein chemistry. We report site specific dual functionalizations of peptides and proteins capitalizing on reactivity differences of cysteines in their free (thiol) and protected, oxidized (disulfide) forms. The dual functionalization of interleukin 2 and EYFP proceeded with no loss of bioactivity in a stepwise fashion applying maleimide and disulfide rebridging allyl-sulfone groups. In order to ensure broader applicability of the functionalization strategy, a novel, short peptide sequence that introduces a disulfide bridge was designed and site-selective dual labeling in the presence of biogenic groups was successfully demonstrated.
Bioconjugate Chemisty
dx.doi.org/10.1021/acs.bioconjchem.7b00675
Single-molecule nucle osome remodeling by INO80 and effects of histone tails
Marianne Schwarz, Kevin Schall, Eleni Kallis, Sebastian Eustermann, Mara Guariento, Manuela Moldt, Karl-Peter Hopfner, Jens Michaelis
Genome maintenance and integrity requires continuous alterations of the compaction state of the chromatin structure. Chromatin remodelers, among others the INO80 complex, help organize chromatin by repositioning, reshaping, or evicting nucleosomes. We report on INO80 nucleosome remodeling, assayed by single‐molecule Foerster resonance energy transfer on canonical nucleosomes as well as nucleosomes assembled from tailless histones. Nucleosome repositioning by INO80 is a processively catalyzed reaction. During the initiation of remodeling, probed by the INO80 bound state, the nucleosome reveals structurally heterogeneous states for tailless nucleosomes (in contrast to wild‐type nucleosomes). We, therefore, propose an altered energy landscape for the INO80‐mediated nucleosome sliding reaction in the absence of histone tails.
FEBS Letters
Copyright © 2018, Federation of European Biochemical Societies, Wiley.
rdcu.be/FYqw

Precision and accuracy in smFRET based structural studies—A benchmark study of the Fast-Nano-Positioning System
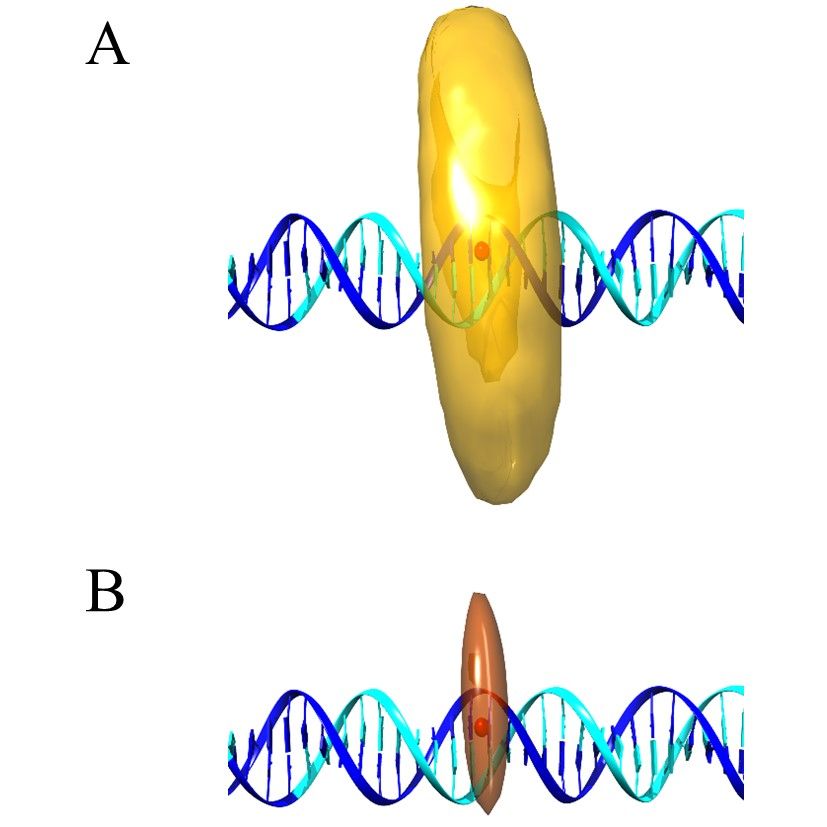
Julia Nagy, Tobias Eilert, Jens Michaelis
Modern hybrid structural analysis methods have opened new possibilities to analyze and resolve flexible protein complexes where conventional crystallographic methods have reached their limits. Here, the Fast-Nano-Positioning System (Fast-NPS), a Bayesian parameter estimation-based analysis method and software, is an interesting method since it allows for the localization of unknown fluorescent dye molecules attached to macromolecular complexes based on single-molecule Förster resonance energy transfer (smFRET) measurements. However, the precision, accuracy, and reliability of structural models derived from results based on such complex calculation schemes are oftentimes difficult to evaluate. Therefore, we present two proof-of-principle benchmark studies where we use smFRET data to localize supposedly unknown positions on a DNA as well as on a protein-nucleic acid complex. Since we use complexes where structural information is available, we can compare Fast-NPS localization to the existing structural data. In particular, we compare different dye models and discuss how both accuracy and precision can be optimized.
The Journal of Chemical Physics
doi.org/10.1063/1.5006477
