Software AG Gebhardt
Analysis of single-molecule tracking data
TrackIt is a tracking and analysis software which allows analyzing and comparing multiple single-molecule fluorescence data sets.
A user-friendly GUI offers convenient tracking visualization, multiple state-of-the-art analysis procedures, display of results, and data im- and export at different levels to utilize external software tools. Furthermore, we developed an algorithm, which accounts for both tracking losses and suggests optimized tracking parameters when evaluating reaction rates.
https://www.nature.com/articles/s41598-021-88802-7
https://gitlab.com/GebhardtLab/TrackIt.
GRID (Genuine Rate IDentification) yields the dissociation rate spectrum of single molecule fluorescence survival time distributions by inverse Laplace transformation and enables correcting for photobleaching by global consideration of all time-lapse conditions. The GRID toolbox also includes a parameter prediction algorithm for nearest neighbour tracking. Parameters predicted by the algorithm ensure homogeneous tracking losses in all time-lapse conditions and thus enable GRID to perform a consistent global fit of all time-lapse conditions.
https://www.nature.com/articles/s41598-020-58634-y
https://gitlab.com/GebhardtLab/GRID
SPLIT (Speedy Partition of Legs In Tracks) is an automatized heuristic to separate different modes of motion (e.g. directed and diffusive motion) within a single track. As a criterion, SPLIT uses spatial information on each position in the track encoded within a recurrence plot. This plot reveals how many detections are in close proximity to each other within a track. Furthermore, SPLIT uses jump angles for classification of directed motion and offers averaging techniques dealing with localization errors in track coordinates.
https://doi.org/10.1101/2020.04.03.023614
https://gitlab.com/GebhardtLab/SPLIT
Analysis of gene regulatory networks
CaiNet (Computer Aided Interactive gene NETwork simulation tool) is a fast computer aided interactive network simulation environment to set up and simulate the temporal evolution of arbitrary gene regulatory networks. CaiNet comes with a graphical user interface that automatically compiles a network into a hybrid stochastic-deterministic simulation framework. For a better understanding of the effects of noise within the network, noise arising from stochastic rates in gene expression can be turned on and off.
https://doi.org/10.1186/s12859-021-04541-6
https://gitlab.com/GebhardtLab/CaiNet
BIRD (Bursting Inference from mRNA Distributions) is an inference algorithm for rate constants involved in bursts of mRNA expression. BIRD uses half analytical solutions for a fast and robust calculation of mRNA distributions from the underlying system of differential equations. To infer rate constants, measured mRNA histograms are fitted with these solutions using gradient-based methods.
https://doi.org/10.1101/2020.11.26.400069
https://gitlab.com/GebhardtLab/BIRD
| Professors: | Prof. Dr. J. Christof M. Gebhardt Prof. Dr. Kay-Eberhard Gottschalk | ||||
| Senior-Professors: | Prof. Dr. sc. nat./ETH Zürich Othmar Marti Prof. Dr. Heinrich Hörber | ||||
| Secretariat: | N25 / 5208
Tel.: +49 (0)731 50 23010 | ||||
| Address: | Institute of Experimental Physics Albert-Einstein-Allee 11 89081 Ulm |
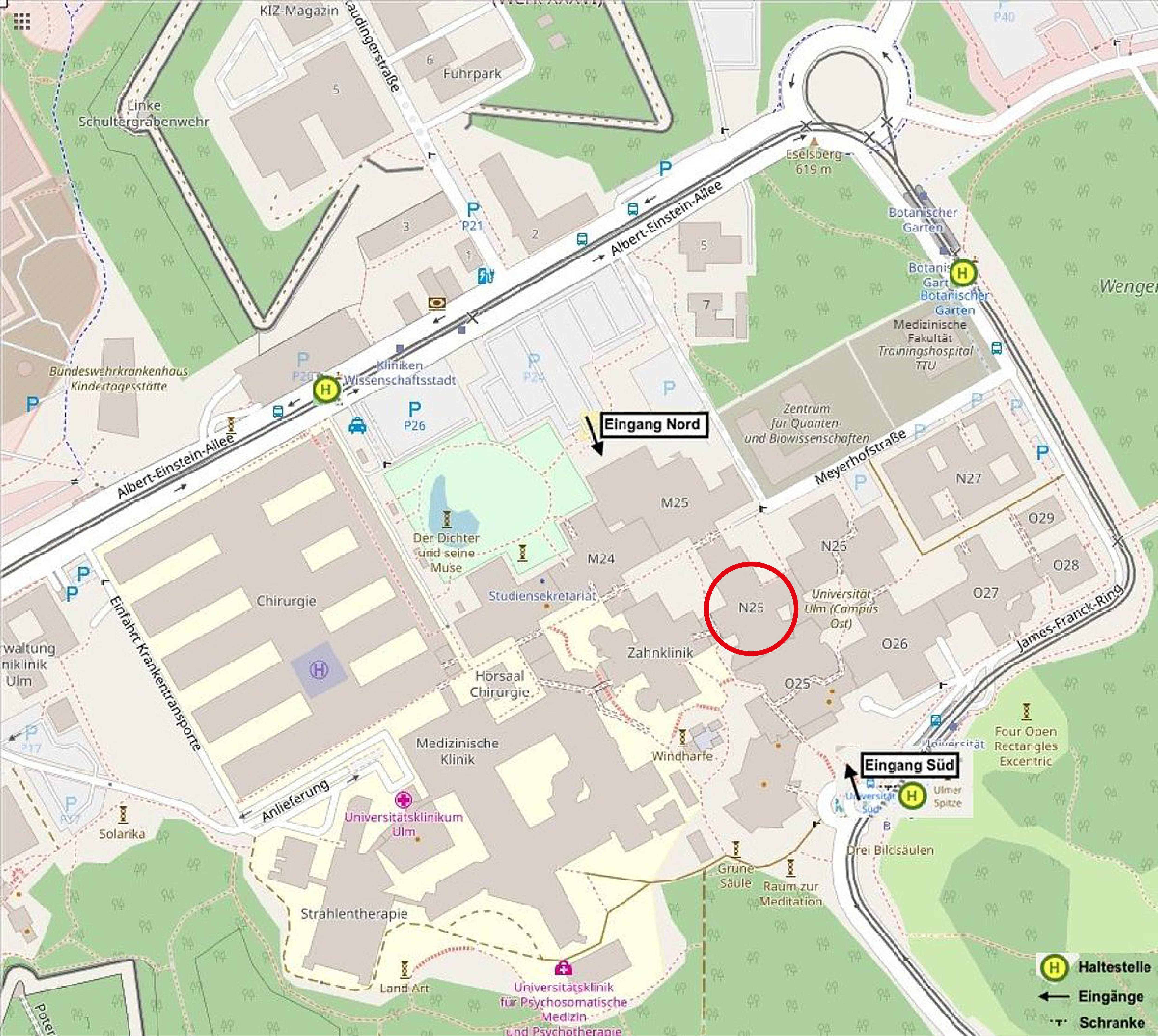
How to reach Ulm University:
Directions, Campusnavigator
e.g. with the tram:
From Ulm main station, every 10 minutes, take tram 2 towards Science Park II until stop Universität Süd.
How to find the institute:
The Institute of Experimental Physics is located in building part N25 of the university East on level 5. The secretariat is in room N25/5208.
from South entrance: go through the entire entry hall towards the North until the staircase/lift of N25.
from North entrance: go left (East) to M25, then right (South) to the staircase/lift of N25.
from East entrance: go left (West) to the staircase/lift of N25.
If you are lost: call the secretariat via 0731 50 23010.
